3.1 Overview
There are three major resolution scales in ST data:
- multi-cell
- single-cell
- sub-cellular
Walker et al. (2022), Yue et al. (2023), Moses & Pachter (2022)
For a practical guide to choosing spatial methods, see Lim et al. (2025).
3.2 Techniques
3.2.1 10X Visium
As of May 2024, there are three assays: Spatial GEX, Spatial GEX + Protein and Spatial GEX HD.
| Assay | Spatial GEX | Spatial GEX+Protein | Spatial GEX HD |
|---|---|---|---|
| Scope | Whole transcriptome | Whole transcriptome+31 Proteins | Whole transcriptome |
| Spatial resolution | multi-cell | multi-cell | single-cell |
| Tissue | FFPE,Fresh frozen,Fixed frozen | FFPE | FFPE |
| Species | Various | Human,Mouse | Human,Mouse |
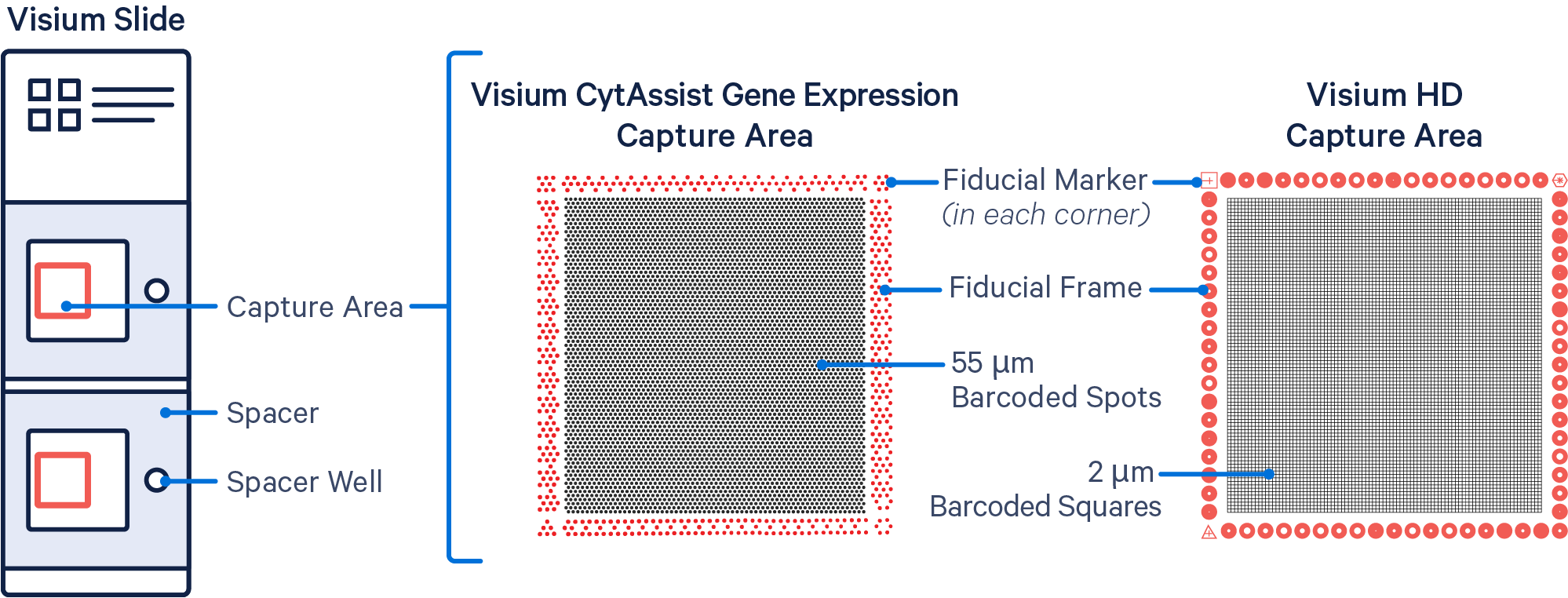
| Capture area | 6.5x6.5mm | 6.5x6.5mm | 6.5x6.5mm |
| Total spots | 5K/14K | 5K/14K | ~11.2M |
| Spot size | 55um circle | 55um circle | 2um square |
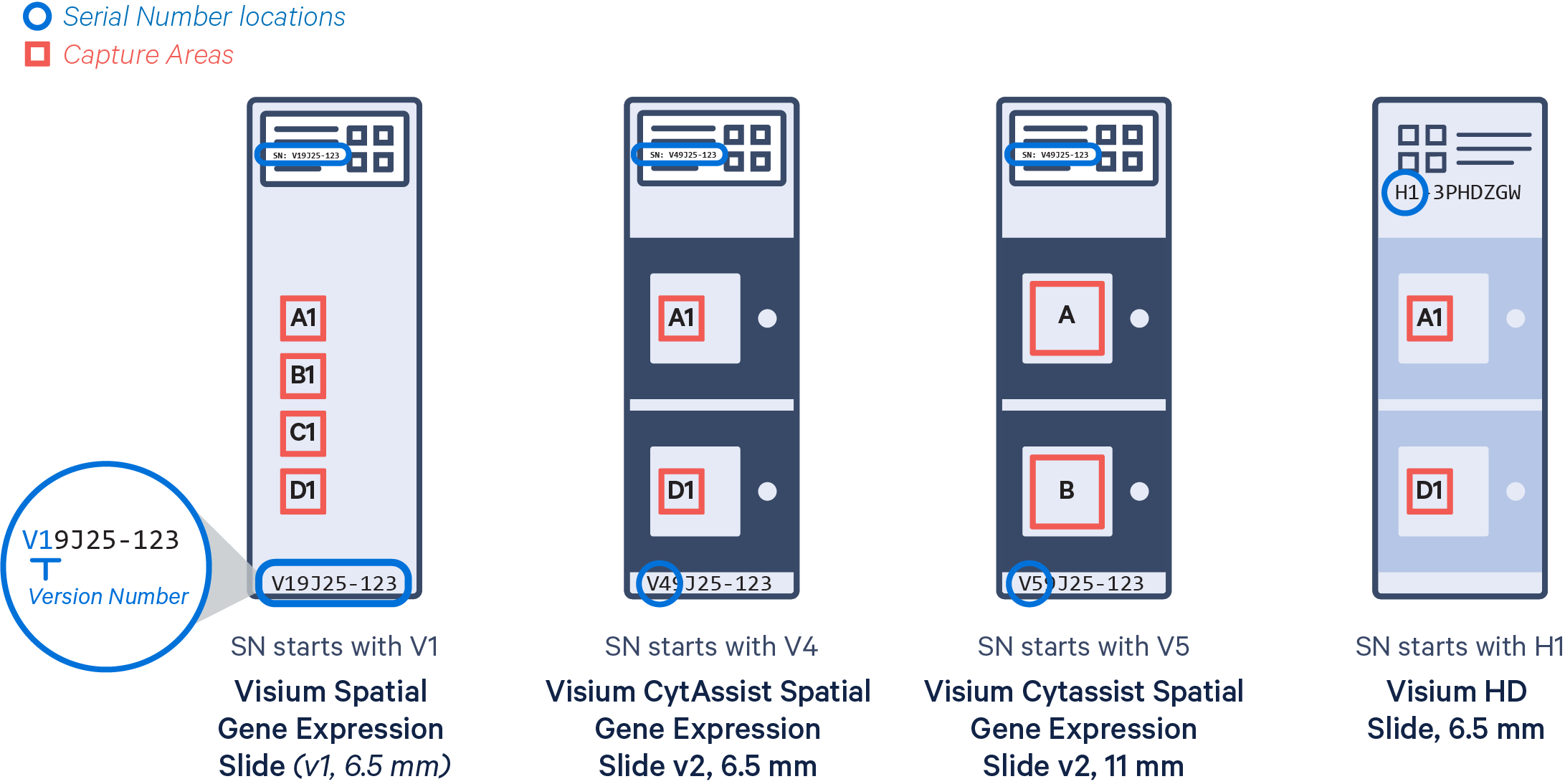
In addition, the Visium slides come in different types depending on the assay and version.
3.3 Methods/Tools
3.3.1 Frameworks
3.3.2 SVG
Identification of spatially variable genes.
- SpatialDE
- SPARK
- SOMDE
- Sepal
- scGCO
- SpaGCN
- SpatialLIBD
- stLearn
3.3.3 Spatial deconvolution
- STdeconvolve (R)
- Reference-free
- RCTD SpaceXR (R)
- Needs reference
- Runs into error
- SPOTlight (R)
- Uses reference
- Giotto SpatialDWLS (R)
- SpatialDDLS (R)
- DSTG (Py)
- Tangram (Py)
3.3.4 Cell interaction
Given the relative stability of cellular locations, spatial transcriptomics allows us to reveal cell–cell interactions (CCI), also referred to as cell-cell communications (CCC), with fewer false positives than similar analysis with scRNA-seq data.
- SpaOTsc
3.4 Datasets
- R package TENxVisiumData
3.5 Resources
References
Lim, H. J., Wang, Y., Buzdin, A., & Li, X. (2025). A practical guide for choosing an optimal spatial transcriptomics technology from seven major commercially available options. BMC Genomics, 26(1), 47. https://doi.org/10.1186/s12864-025-11235-3
Moses, L., & Pachter, L. (2022). Museum of spatial transcriptomics. Nature Methods, 19(5), 534–546. https://www.nature.com/articles/s41592-022-01409-2
Walker, B. L., Cang, Z., Ren, H., Bourgain-Chang, E., & Nie, Q. (2022). Deciphering tissue structure and function using spatial transcriptomics. Communications Biology, 5(1), 220.
Yue, L., Liu, F., Hu, J., Yang, P., Wang, Y., Dong, J., Shu, W., Huang, X., & Wang, S. (2023). A guidebook of spatial transcriptomic technologies, data resources and analysis approaches. Computational and Structural Biotechnology Journal. https://www.sciencedirect.com/science/article/pii/S2001037023000156