Bioinformatics Data Analysis
| 2018 - Present | Bioinformatician | National Bioinformatics Infrastructure Sweden, Uppsala, Sweden |
I current perform bioinformatics data analyses of high throughput sequencing data originating from DNA or RNA-Seq data in addition to methylation and expression microarrays.
Germline Transcriptomics in Zebrafish

| 2014 - 2018 | Post-Doctoral researcher | Uppsala University, Uppsala, Sweden |
My post-doctoral research work explored the consequences of haploid selection in Zebrafish. Zebrafish sperm were artificially selected through in-vitro fertilisation for specific traits and the following three generations were monitored for phenotypic and genetic changes. The DNA and RNA were extracted and analysed using high throughput sequencing to understand changes at the molecular level.
Honey Bee Research

| 2009 - 2013 | PhD / Post-Doctoral researcher | Aarhus University, Slagelse, Denmark |
I worked on European honey bees ( Apis mellifera ) as part of my PhD work. This included viral diseases, diagnosis and quantification. I was also involved in honey bee health and vitality as part of an EU project. I also worked on the population structure and spatial distribution of honey bee subspecies in Europe.
Characterisation of Chitinase genes from bacterium Pantoea dispersa
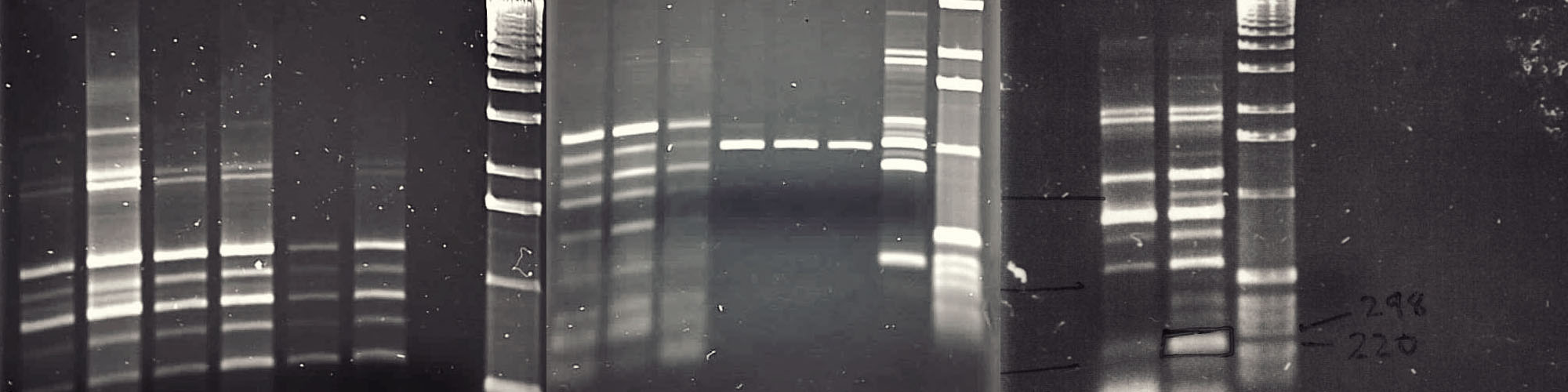
| 2008 - 2009 | Research assistant | University of Hertfordshire, Hatfield, UK |
Chitin is a biopolymer found in many organisms such as arthropods and insects as well as in plants and fungi. Chitinases are enzymes that break down chitin. Chitinases are produced by a wide range of organisms, and their composition and functional properties may differ. In nature, chitin is part of the exoskeleton in insects, the cell wall of fungi or even a defensive mechanism in plants. Chitin and chitinases have extensive use in agriculture and industry including medical applications, food processing, fertilizer and nanomaterial research. Therefore, it is important to study the activity and stability of these enzymes using molecular approaches. A novel of class of chitinase genes were discovered in bacterium Pantoea dispersa. This project involved bacterial culture followed by identification, amplification, sequencing and sequence comparison of the chitinase genes in this bacteria.
Lux-integrated bioluminescent Pseudomonas aeruginosa as a biosensor
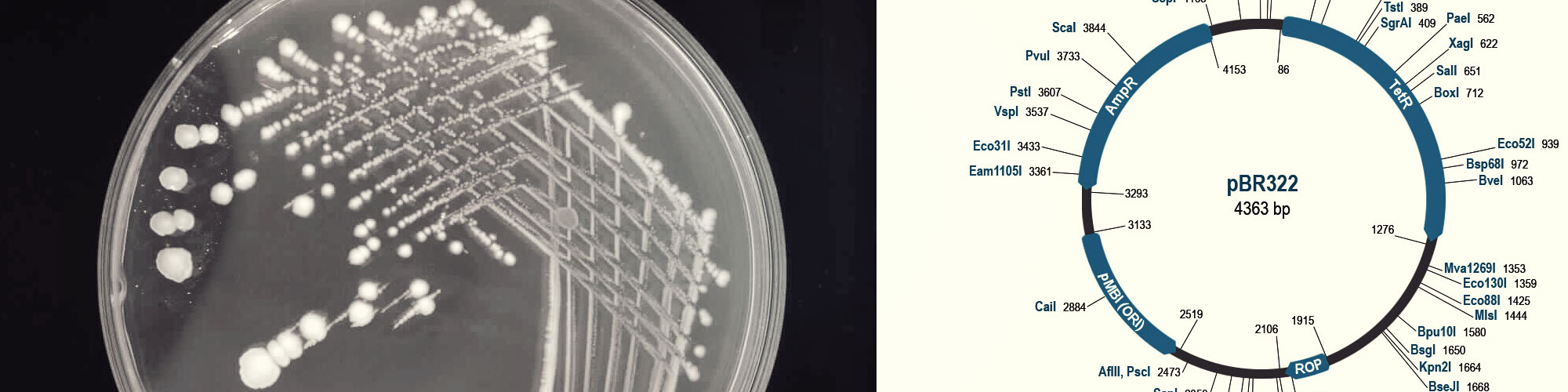
| 2007 - 2008 | Master’s Project | University of Hertfordshire, Hatfield, UK |
Microbes such as bacteria can be used as biosensors for a range of agricultural, environmental and industrial applications from monitoring toxicity in water bodies, detection of pathogens and contamination in industrial effluents. One particular application is the testing of pharmaceutical products for microbial activity. The traditional approach using viable count technique is time consuming, expensive and laborious. An alternative approach to bacterial quantification is proposed using measurement of light intensity in the samples using bioluminescent bacteria. This project involved the transformation of bacterium Pseudomonas aeruginosa with a pBR322 plasmid containing the bioluminescent lux gene construct.
