This is an update to an older post from 2015 on the same topic. This covers the exact same thing but using the latest R packages and coding style using the pipes (%>% ) and tidyverse packages.
This was inspired by the disease incidence rate in the US featured on the Wall Street Journal. The disease incidence dataset was originally used in this article in the New England Journal of Medicine. In this post, I am using the measles level 1 incidence (cases per 100,000 people) dataset obtained as a .csv file from Project Tycho. Download the .csv file here.
In this post, we will look into creating a neat, clean and elegant heatmap in R. No clustering, no dendrograms, no trace lines, no bullshit. We will go through some basic data cleanup, reformatting and finally plotting. We go through this step by step. For the whole code with minimal explanations, scroll to the bottom of the page.
I am running R version 3.5.2 64-bit on Ubuntu 18.04 64-bit. Packages in use are ggplot2 (3.1.0), dplyr (0.7.8), tidyr (0.8.2), stringr (1.3.1) and for base plots I am using gplots (3.0.1.1) and plotrix (3.7-4). Install the necessary packages if not already installed and load them.
# install packages
# install.packages(pkgs = c("ggplot2","dplyr","tidyr","stringr","gplots","plotrix"),dependencies = T)
# load packages
library(ggplot2) # ggplot() for plotting
library(dplyr) # data reformatting
library(tidyr) # data reformatting
library(stringr) # string manipulation
1. Data preparation
Read in the .csv file and inspect the data. The first two rows with non-table data is skipped.
# read csv file
m <- read.csv("measles_lev1.csv",header=T,stringsAsFactors=F,skip=2)
# inspect data
head(m)
str(m)
table(m$YEAR)
table(m$WEEK)
The head() function shows us the header names and the first 6 rows of the data. The str() function shows that YEAR and WEEK columns are stored as integers and the incidences as characters. The incidences although numeric have been read in as character because missing values are coded as “-“. The table() function is used to check for any missing years or weeks. The data is currently stored in so called ‘wide’ format which we will convert to ‘long’ format. The ggplot2 plotting package prefers long format. The wide format is shown below.
> head(m)
YEAR WEEK ALABAMA ALASKA ARIZONA ARKANSAS CALIFORNIA COLORADO
1 1928 1 3.67 - 1.90 4.11 1.38 8.38
2 1928 2 6.25 - 6.40 9.91 1.80 6.02
3 1928 3 7.95 - 4.50 11.15 1.31 2.86
4 1928 4 12.58 - 1.90 13.75 1.87 13.71
5 1928 5 8.03 - 0.47 20.79 2.38 5.13
6 1928 6 7.27 - 6.40 26.58 2.79 8.09
The YEARand WEEK variables are kept as is and all incidence values are collapsed into a variable and value column. The column names are changed to lower case for convenience. The year and week variables are converted to factors and value variable is converted to a number.
m2 <- m %>%
# convert data to long format
gather(key="state", value="value", -YEAR, -WEEK) %>%
# rename columns
setNames(c("year", "week", "state", "value")) %>%
# convert year to factor
mutate(year=factor(year)) %>%
# convert week to factor
mutate(week=factor(week)) %>%
# convert value to numeric (also converts '-' to NA, gives a warning)
mutate(value=as.numeric(value))
The output in long format is shown below.
> head(m2)
year week state value
1 1928 1 Alabama 3.67
2 1928 2 Alabama 6.25
3 1928 3 Alabama 7.95
4 1928 4 Alabama 12.58
5 1928 5 Alabama 8.03
6 1928 6 Alabama 7.27
The state variable is currently in all caps and multi-word states have dot separators. I prefer to have them in Title Case and separated by space to be shown on the plot later. A custom function is used to change the states into Title Case. Multi-word states are split at the dot separator and each word is changed into Title Case using the function str_to_title() and they are pasted back together.
# removes . and change states to title case using custom function
fn_tc <- function(x) paste(str_to_title(unlist(strsplit(x, "[.]"))), collapse=" ")
m2$state <- sapply(m2$state, fn_tc)
Now, I would like to plot the heatmap with the year on the x-axis and state on the y-axis, which means we have to deal with the week variable in some way. We will sum all the incidences from all weeks for each year and discard the week variable. The dplyr compliant way to do this is to use group_by() followed by summarise() using a function.
The way sum() handles NAs is a bit strange. By default, it returns NA if one or more elements in the input vector is NA. If we set argument na.rm=TRUE, then NAs are removed and the remaining numbers are summed. But if all the elements are NA, the sum is returned as zero. This is weird and undesirable in this situation. Therefore, I have a custom sum function named na_sum() to remove NAs and return the remaining sum or return NA if all elements are NA. We then use this custom function inside summarise() function to summarise the data by year and state while getting rid of week.
# custom sum function returns NA when all values in set are NA,
# in a set mixed with NAs, NAs are removed and remaining summed.
na_sum <- function(x)
{
if(all(is.na(x))) val <- sum(x, na.rm=F)
if(!all(is.na(x))) val <- sum(x, na.rm=T)
return(val)
}
# sum incidences for all weeks into one year
m3 <- m2 %>%
group_by(year, state) %>%
summarise(count=na_sum(value)) %>%
as.data.frame()
Now our data looks like this without the week variable. The values are summed by year for each state to create a new variable count.
> head(m3)
year state count
1 1928 Alabama 334.99
2 1928 Alaska 0.00
3 1928 Arizona 200.75
4 1928 Arkansas 481.77
5 1928 California 69.22
6 1928 Colorado 206.98
At this point, the data preparation is essentially over. The data is in ‘long’ format, the x, y and z variables for the plot are available and are of the correct type: factor, factor and numeric. If your data is already in this format, it is easy to jump straight into visualisation. But depending on what sort of data you start off with, the data preparation and reformatting can be complicated and tedious.
2. Plotting
I prefer to use the ggplot2 plotting package to plot graphs in R due to its consistent code structure. I will focus mostly on ggplot2 code here. But, just for the sake of completeness, I will also include some heatmap code using base graphics.
2.1 ggplot2
Once the data is in the right format, plotting the data is rather simple code in ggplot2. The ggplot2 index page has the code syntax and arguments.
#basic ggplot
p <- ggplot(m3, aes(x=year, y=state, fill=count))+
geom_tile()
#save plot to working directory
ggsave(p, filename="measles-basic.png")
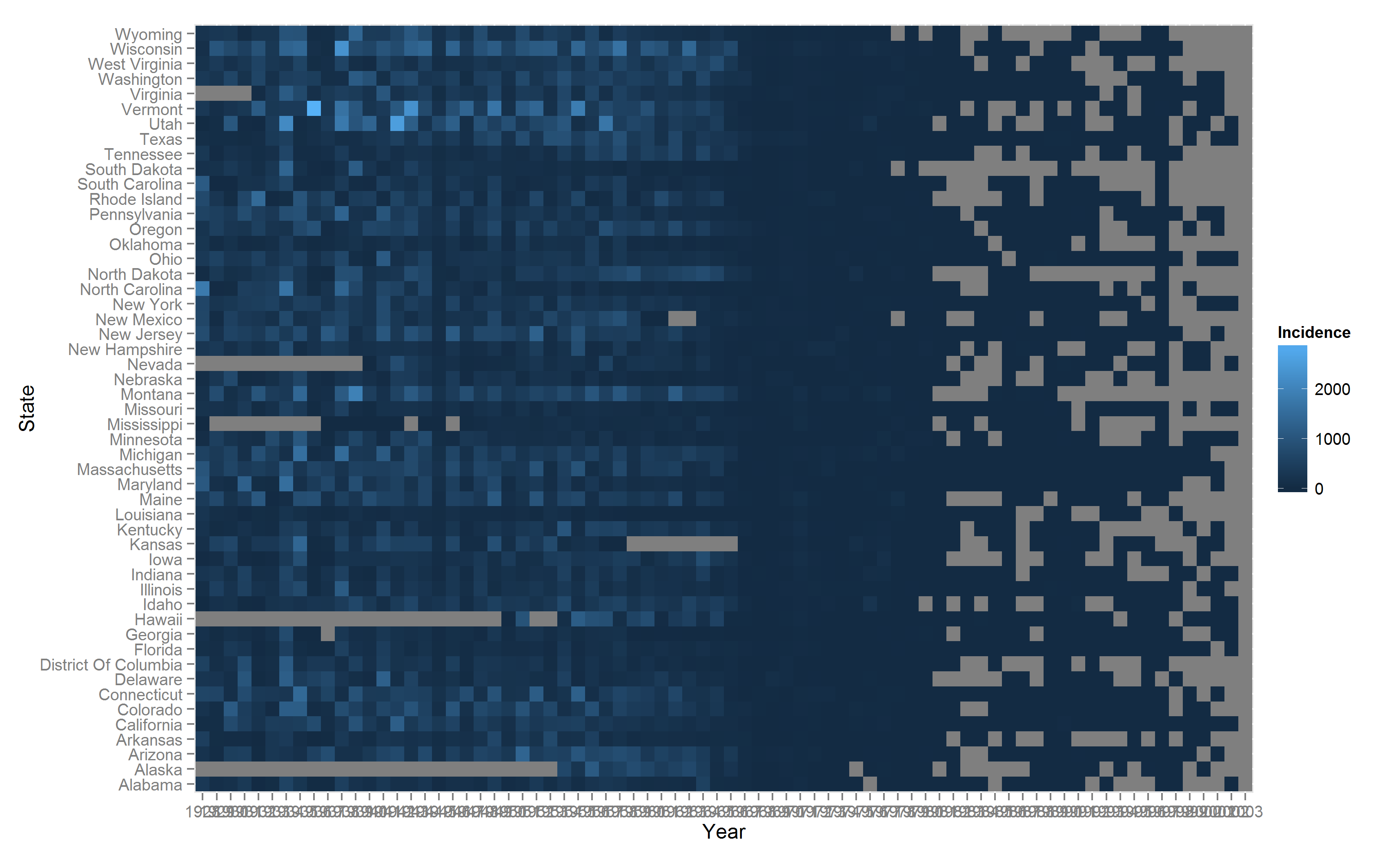
The default output from ggplot2 is quite decent- There are several aspects of the basic plot that can be modified and tweaked. We add tile borders, custom x-axis breaks and custom text sizes. The ggplot code is modified below.
#modified ggplot
p <- ggplot(m3, aes(x=year, y=state, fill=count))+
#add border white colour of line thickness 0.25
geom_tile(colour="white", size=0.25)+
#remove x and y axis labels
labs(x="", y="")+
#remove extra space
scale_y_discrete(expand=c(0, 0))+
#define new breaks on x-axis
scale_x_discrete(expand=c(0, 0),
breaks=c("1930", "1940", "1950", "1960", "1970", "1980", "1990", "2000"))+
#set a base size for all fonts
theme_grey(base_size=8)+
#theme options
theme(
#bold font for legend text
legend.text=element_text(face="bold"),
#set thickness of axis ticks
axis.ticks=element_line(size=0.4),
#remove plot background
plot.background=element_blank(),
#remove plot border
panel.border=element_blank()
)
#save with dpi 200
ggsave(p, filename="measles-mod1.png", height=5.5, width=8.8, units="in", dpi=200)
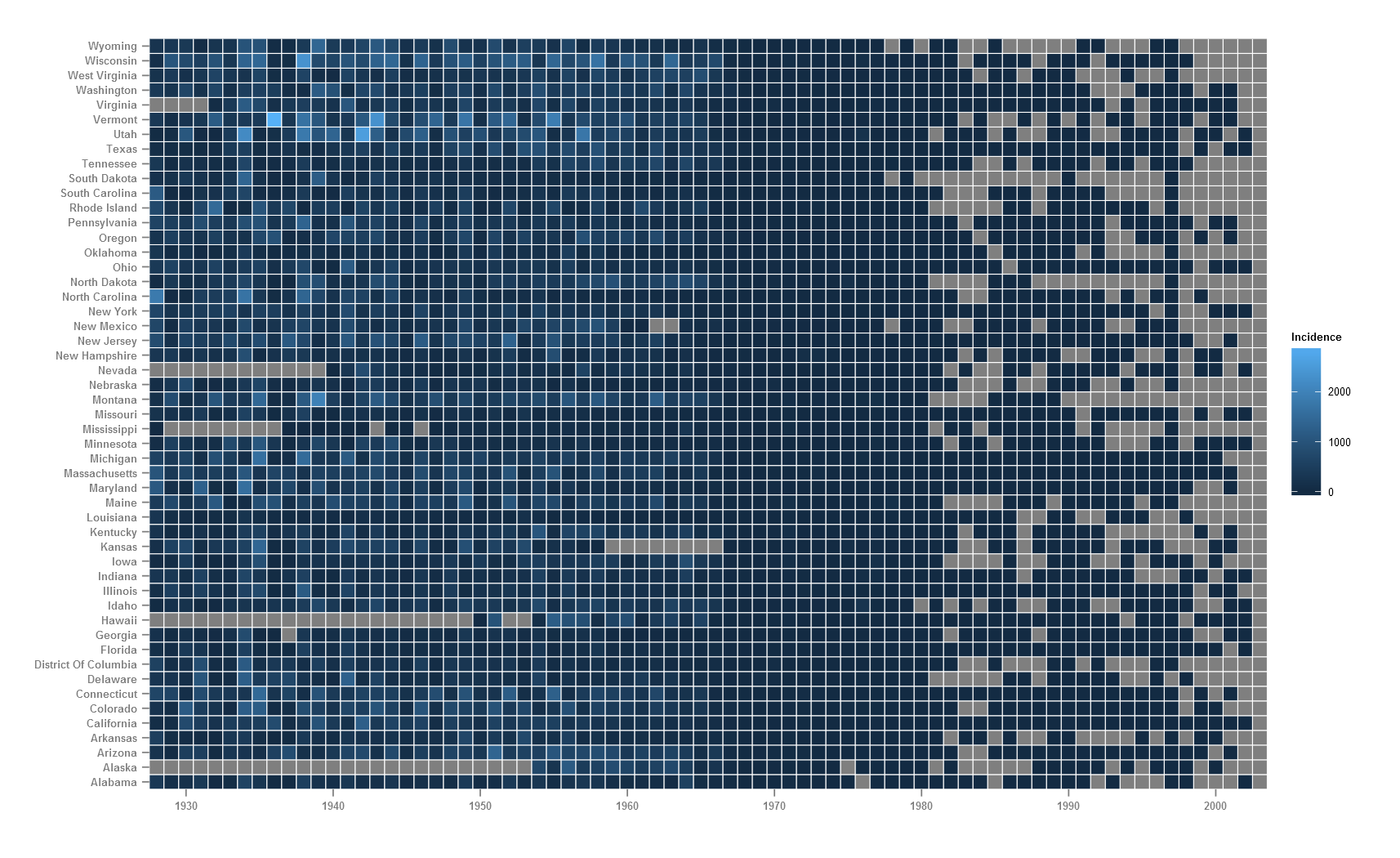
I would prefer the y-axis labels (states) to be ordered ascending top-bottom. This means going back to our ‘long’ format data and refactoring the state variable in reverse. The fill variable here (count) is a continuous variable and hence, ggplot by default uses the blue colour ramp. Here and in many cases, it might make better sense to bin the continuous data into levels and represent each level as a discrete colour. The cut() function in R allows to break and label a continuous variable.
The count variable is binned into 7 levels and saved as a new variable countfactor. The NAs remain as NA. Defining breaks in your variable depends on the type of data, the number of bins that make sense with the context or just trial and error. Too many bins are not good. Checking your variable using summary(x) or a boxplot(x) can reveal a lot about the data.
m4 <- m3 %>%
# convert state to factor and reverse order of levels
mutate(state=factor(state, levels=rev(sort(unique(state))))) %>%
# create a new variable from count
mutate(countfactor=cut(count, breaks=c(-1, 0, 1, 10, 100, 500, 1000, max(count, na.rm=T)),
labels=c("0", "0-1", "1-10", "10-100", "100-500", "500-1000", ">1000"))) %>%
# change level order
mutate(countfactor=factor(as.character(countfactor), levels=rev(levels(countfactor))))
Now we are ready to plot the final dataset.
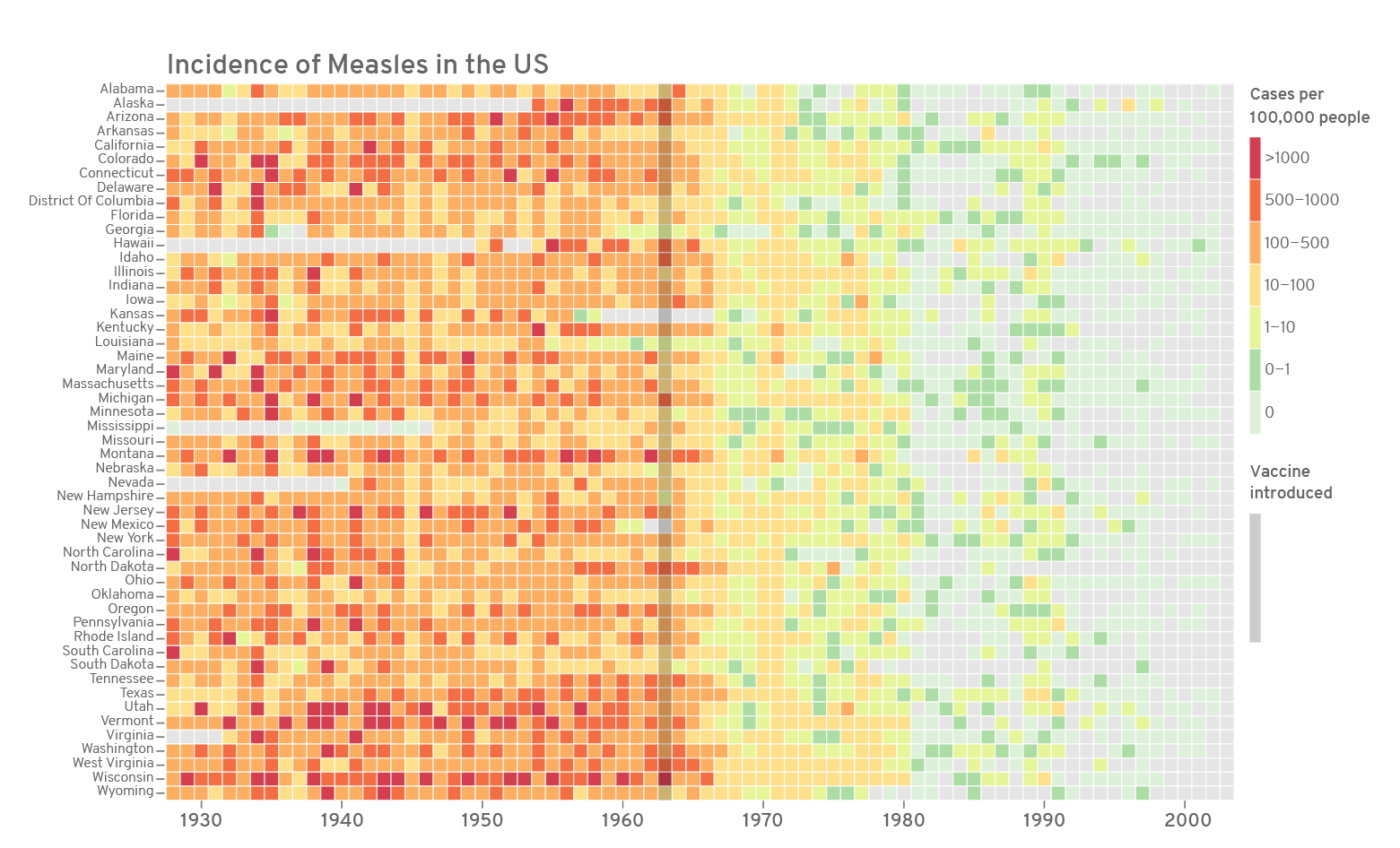
# assign text colour
textcol <- "grey40"
# further modified ggplot
p <- ggplot(m4, aes(x=year, y=state, fill=countfactor))+
geom_tile(colour="white", size=0.2)+
guides(fill=guide_legend(title="Cases per\n100,000 people"))+
labs(x="", y="", title="Incidence of Measles in the US")+
scale_y_discrete(expand=c(0, 0))+
scale_x_discrete(expand=c(0, 0), breaks=c("1930", "1940", "1950", "1960", "1970", "1980", "1990", "2000"))+
scale_fill_manual(values=c("#d53e4f", "#f46d43", "#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da"), na.value = "grey90")+
#coord_fixed()+
theme_grey(base_size=10)+
theme(legend.position="right", legend.direction="vertical",
legend.title=element_text(colour=textcol),
legend.margin=margin(grid::unit(0, "cm")),
legend.text=element_text(colour=textcol, size=7, face="bold"),
legend.key.height=grid::unit(0.8, "cm"),
legend.key.width=grid::unit(0.2, "cm"),
axis.text.x=element_text(size=10, colour=textcol),
axis.text.y=element_text(vjust=0.2, colour=textcol),
axis.ticks=element_line(size=0.4),
plot.background=element_blank(),
panel.border=element_blank(),
plot.margin=margin(0.7, 0.4, 0.1, 0.2, "cm"),
plot.title=element_text(colour=textcol, hjust=0, size=14, face="bold")
)
#export figure
ggsave(p, filename="measles-mod3.png", height=5.5, width=8.8, units="in", dpi=200)
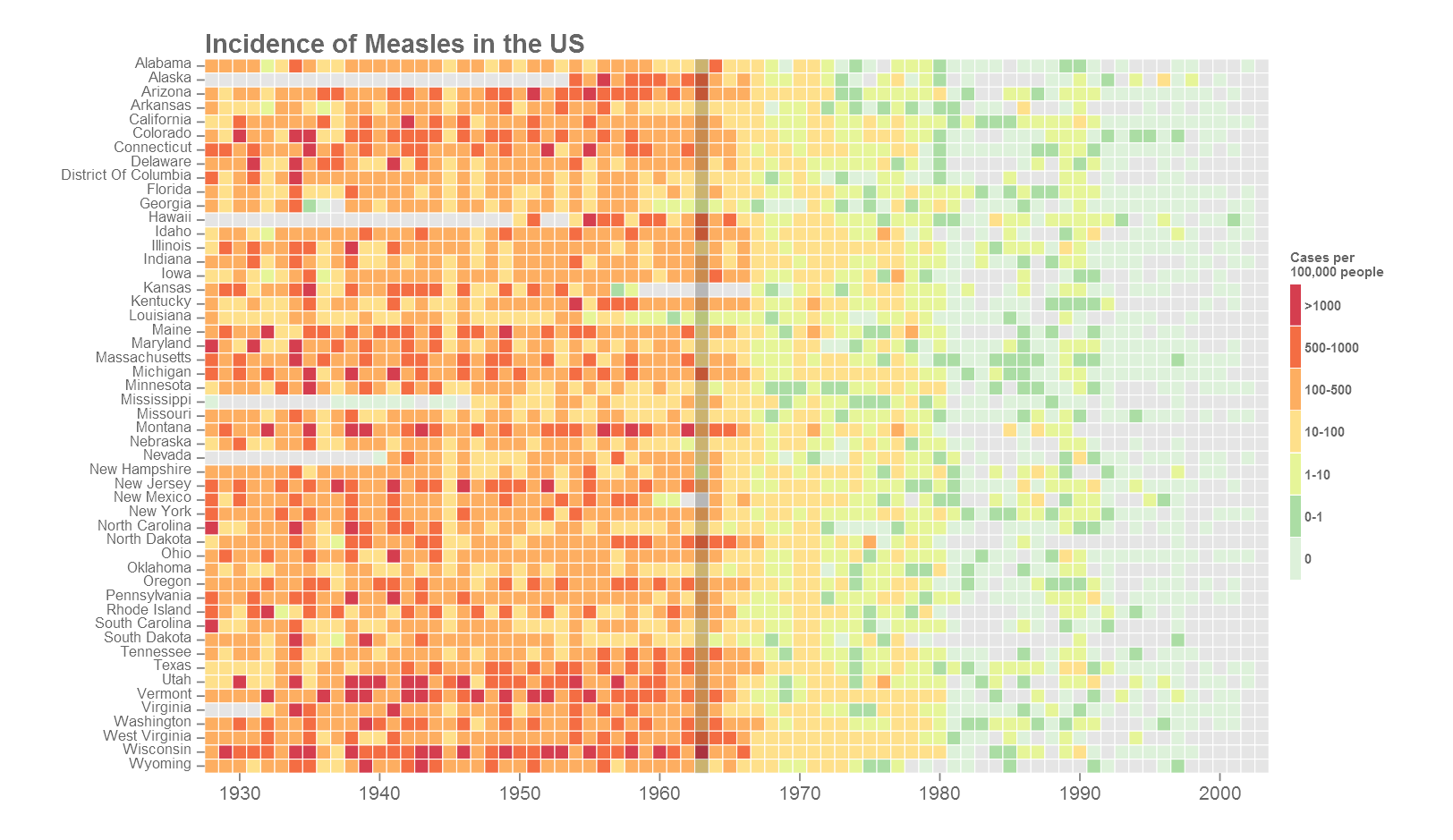
This is the final version of the figure. All font elements are coloured grey40. Missing values (NAs) are coloured grey90. The year of the introduction of vaccination is indicated as a dark vertical stripe. If the y-axis labels are too many or too small, they can be dropped the same way as the x-axis. A custom colour palette was used from ColorBrewer based on the Spectral palette. Using the R package RColorBrewer() or using ggplot2 function scale_fill_brewer() opens up all the colour palettes from the ColorBrewer website. Here is an example below:
library(RColorBrewer)
#change the scale_fill_manual from previous code to below
scale_fill_manual(values=rev(brewer.pal(7, "YlGnBu")), na.value="grey90")+
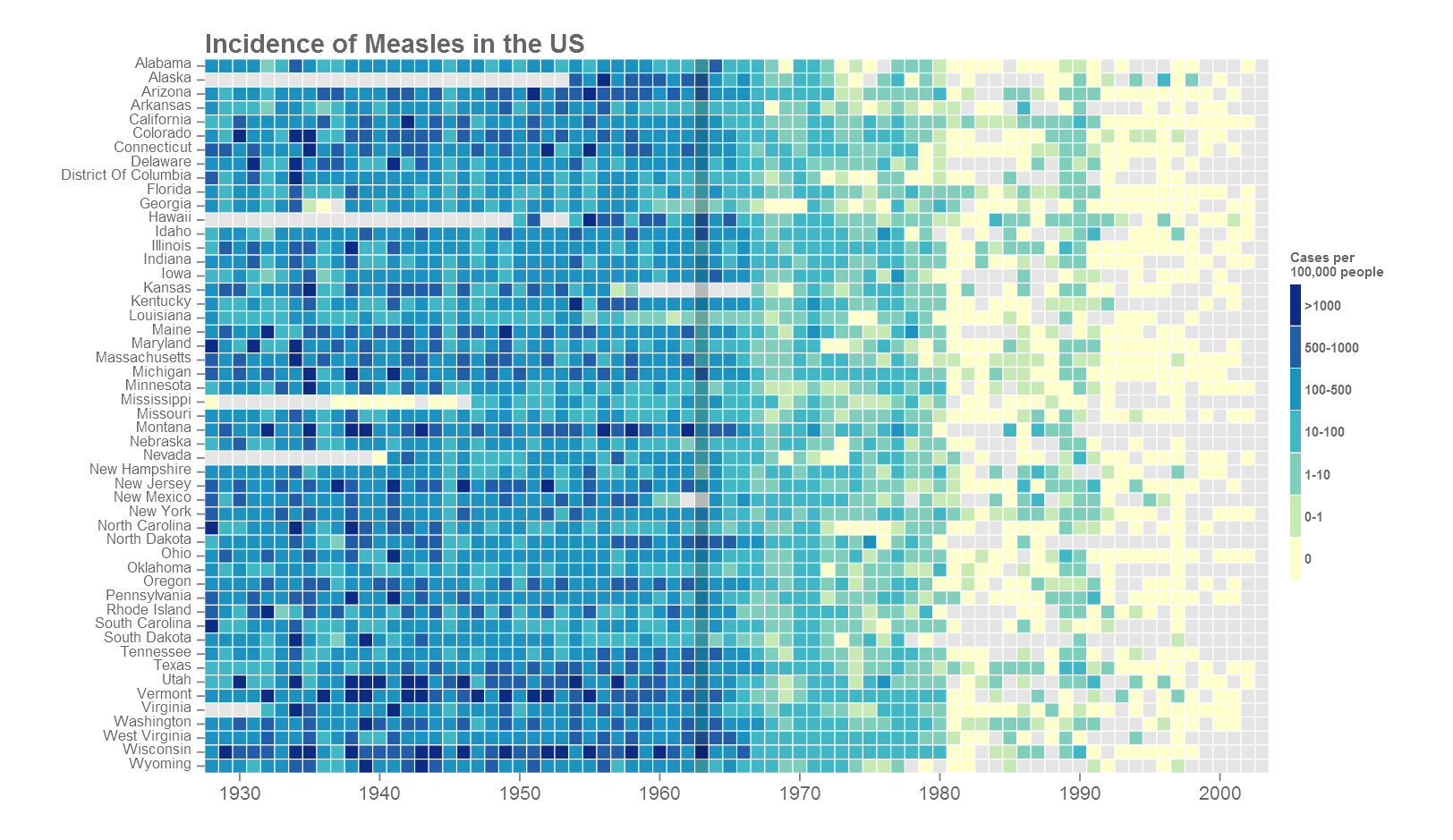
Most of the plot customisation happens in the theme() section of the code. Refer to the ggplot index for all theme arguments. Based on where the figure will go next, the fonts may need to be changed. The extrafont() package comes handy. Another option is to export in vector format such as .svg or .pdf. Import into a vector editor such as Adobe Illustrator and add your own text. This will produce nice crisp results but does take some manual work. See the cover image of this post for example.
2.2 Base graphics
We will take a quick look at plotting using base graphics. The base function heatmap() and the enchanced heatmap.2() function from the gplots package uses a wide format data matrix as input. We start with the ‘long’ data that we prepared in section 1. We convert the data in ‘long’ format into ‘wide’ format using the spread() function from package tidyr. The wide data is converted to a matrix after removal of non-numeric columns. The state names are reassigned as rownames of the matrix to be used as y-axis text.
# load package
library(gplots) # heatmap.2() function
library(plotrix) # gradient.rect() function
# convert from long format to wide format
m5 <- m3 %>% spread(key="state", value=count)
m6 <- as.matrix(m5[ ,-1])
rownames(m6) <- m5$year
#base heatmap
png(filename="measles-base.png", height=5.5, width=8.8, res=200, units="in")
heatmap(t(m6), Rowv=NA, Colv=NA, na.rm=T, scale="none", col=terrain.colors(100),
xlab="", ylab="", main="Incidence of Measles in the US")
dev.off()
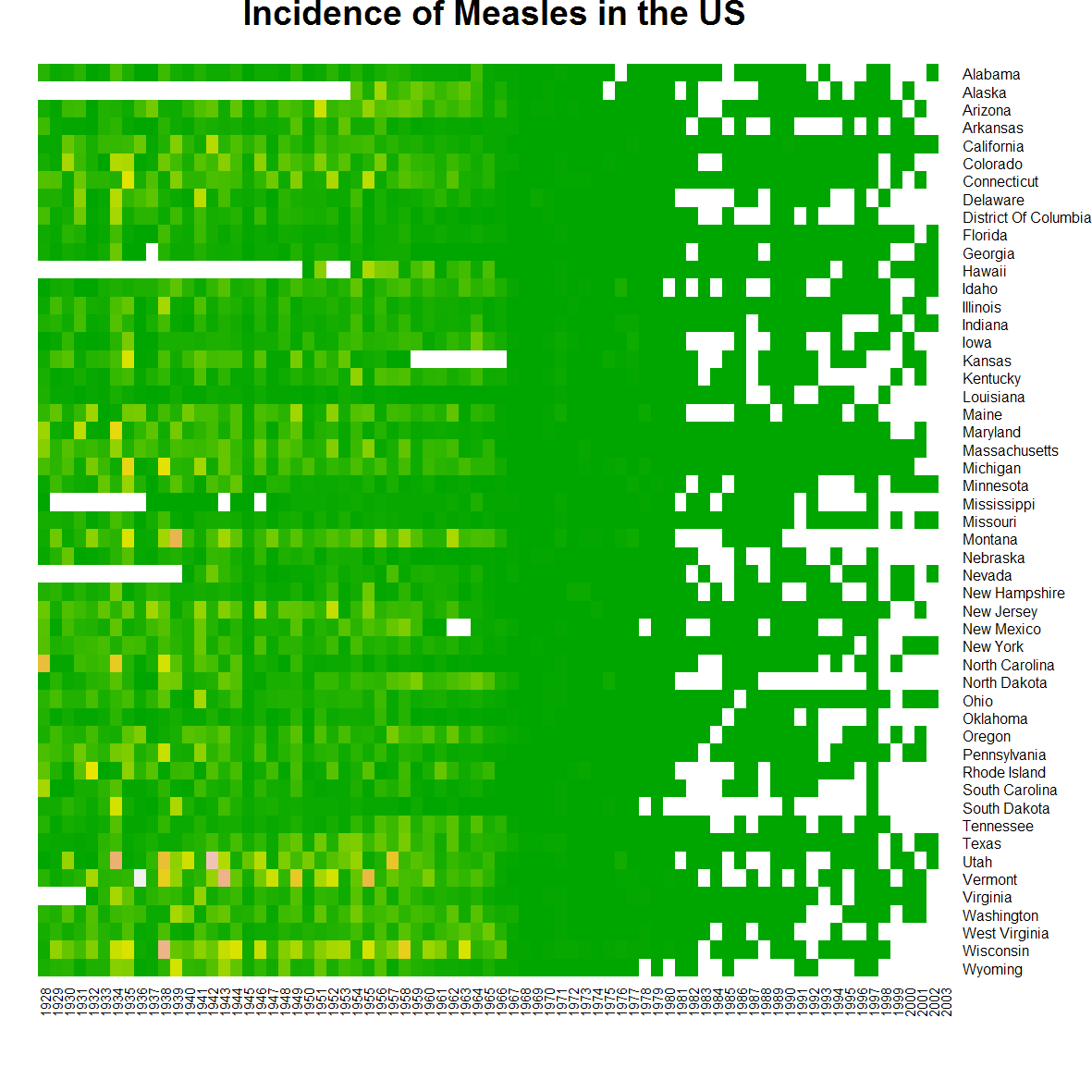
heatmap().
That’s pretty much what we can do with that. The enchanced heatmap.2() function from gplots package can do more. The legend for the colours is not the best, so we will use the function gradient.rect() from plotrix package to create our own legend. And after fiddling around with numerous arguments and countless trial and errors later, we have as below.
# gplots heatmap.2
png(filename="measles-gplot.png", height=6, width=9, res=200, units="in")
par(mar=c(2, 3, 3, 2))
gplots::heatmap.2(t(m6), na.rm=T, dendrogram="none", Rowv=NULL, Colv="Rowv", trace="none", scale="none", offsetRow=0.3, offsetCol=0.3, breaks=c(-1, 0, 1, 10, 100, 500, 1000, max(m4$count, na.rm=T)), colsep=which(seq(1928, 2003)%%10==0), margin=c(3, 8), col=rev(c("#d53e4f", "#f46d43", "#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da")),
xlab="", ylab="", key=F, lhei=c(0.1, 0.9), lwid=c(0.2, 0.8))
gradient.rect(0.125, 0.25, 0.135, 0.75, nslices=7, border=F, gradient="y", col=rev(c("#d53e4f", "#f46d43", "#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da")))
text(x=rep(0.118, 7), y=seq(0.28, 0.72, by=0.07), adj=1, cex=0.8, labels=c("0", "0-1", "1-10", "10-100", "100-500", "500-1000", ">1000"))
text(x=0.135, y=0.82, labels="Cases per\n100,000 people", adj=1, cex=0.85)
title(main="Incidence of Measles in the US", line=1, oma=T, adj=0.21)
dev.off()
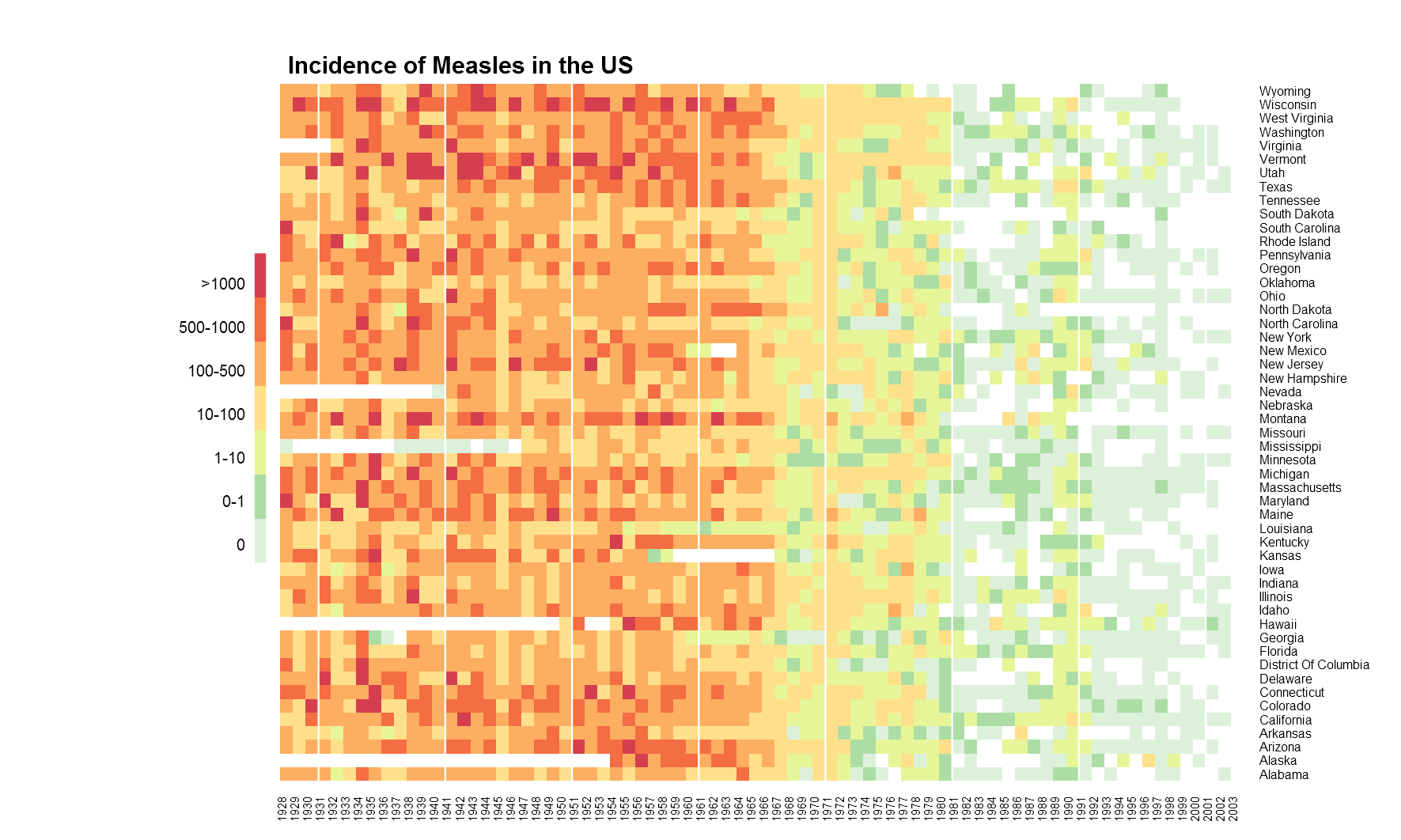
heatmap.2().
Ultimately, we get something useful. The white separator line is a neat feature. It can be used to group columns or rows as required. The while vertical lines above group decades. And below is the full undisrupted code.
# 2019 | Roy Mathew Francis
# Heatmap R code
#load packages
library(ggplot2) # ggplot() for plotting
library(dplyr) # data reformatting
library(tidyr) # data reformatting
library(stringr) # string manipulation
# DATA PREPARATION -------------------------------------------------------------
#read csv file
m <- read.csv("measles_lev1.csv", header=T, stringsAsFactors=F, skip=2)
m2 <- m %>%
# convert data to long format
gather(key="state", value="value", -YEAR, -WEEK) %>%
# rename columns
setNames(c("year", "week", "state", "value")) %>%
# convert year to factor
mutate(year=factor(year)) %>%
# convert week to factor
mutate(week=factor(week)) %>%
# convert value to numeric (also converts '-' to NA, gives a warning)
mutate(value=as.numeric(value))
# removes . and change states to title case using custom function
fn_tc <- function(x) paste(str_to_title(unlist(strsplit(x, "[.]"))), collapse=" ")
m2$state <- sapply(m2$state, fn_tc)
# custom sum function returns NA when all values in set are NA,
# in a set mixed with NAs, NAs are removed and remaining summed.
na_sum <- function(x)
{
if(all(is.na(x))) val <- sum(x, na.rm=F)
if(!all(is.na(x))) val <- sum(x, na.rm=T)
return(val)
}
# sum incidences for all weeks into one year
m3 <- m2 %>%
group_by(year, state) %>%
summarise(count=na_sum(value)) %>%
as.data.frame()
m4 <- m3 %>%
# convert state to factor and reverse order of levels
mutate(state=factor(state, levels=rev(sort(unique(state))))) %>%
# create a new variable from count
mutate(countfactor=cut(count, breaks=c(-1, 0, 1, 10, 100, 500, 1000, max(count, na.rm=T)),
labels=c("0", "0-1", "1-10", "10-100", "100-500", "500-1000", ">1000"))) %>%
# change level order
mutate(countfactor=factor(as.character(countfactor), levels=rev(levels(countfactor))))
# GGPLOT -----------------------------------------------------------------------
# assign text colour
textcol <- "grey40"
# further modified ggplot
p <- ggplot(m4, aes(x=year, y=state, fill=countfactor))+
geom_tile(colour="white", size=0.2)+
guides(fill=guide_legend(title="Cases per\n100,000 people"))+
labs(x="", y="", title="Incidence of Measles in the US")+
scale_y_discrete(expand=c(0, 0))+
scale_x_discrete(expand=c(0, 0), breaks=c("1930", "1940", "1950", "1960", "1970", "1980", "1990", "2000"))+
scale_fill_manual(values=c("#d53e4f", "#f46d43", "#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da"), na.value = "grey90")+
#coord_fixed()+
theme_grey(base_size=10)+
theme(legend.position="right", legend.direction="vertical",
legend.title=element_text(colour=textcol),
legend.margin=margin(grid::unit(0, "cm")),
legend.text=element_text(colour=textcol, size=7, face="bold"),
legend.key.height=grid::unit(0.8, "cm"),
legend.key.width=grid::unit(0.2, "cm"),
axis.text.x=element_text(size=10, colour=textcol),
axis.text.y=element_text(vjust=0.2, colour=textcol),
axis.ticks=element_line(size=0.4),
plot.background=element_blank(),
panel.border=element_blank(),
plot.margin=margin(0.7, 0.4, 0.1, 0.2, "cm"),
plot.title=element_text(colour=textcol, hjust=0, size=14, face="bold")
)
# export figure
ggsave(p, filename="measles-mod3.png", height=5.5, width=8.8, units="in", dpi=200)
# BASE GRAPHICS ----------------------------------------------------------------
# load package
library(gplots) # heatmap.2() function
library(plotrix) # gradient.rect() function
# convert from long format to wide format
m5 <- m3 %>% spread(key="state", value=count)
m6 <- as.matrix(m5[ ,-1])
rownames(m6) <- m5$year
# base heatmap
png(filename="measles-base.png", height=5.5, width=8.8, res=200, units="in")
heatmap(t(m6), Rowv=NA, Colv=NA, na.rm=T, scale="none", col=terrain.colors(100),
xlab="", ylab="", main="Incidence of Measles in the US")
dev.off()
# gplots heatmap.2
png(filename="measles-gplot.png", height=6, width=9, res=200, units="in")
par(mar=c(2, 3, 3, 2))
gplots::heatmap.2(t(m6), na.rm=T, dendrogram="none", Rowv=NULL, Colv="Rowv", trace="none", scale="none",offsetRow=0.3, offsetCol=0.3, breaks=c(-1, 0, 1, 10, 100, 500, 1000, max(m4$count, na.rm=T)), colsep=which(seq(1928, 2003)%%10==0), margin=c(3, 8), col=rev(c("#d53e4f", "#f46d43", "#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da")), xlab="", ylab="", key=F, lhei=c(0.1, 0.9), lwid=c(0.2, 0.8))
gradient.rect(0.125, 0.25, 0.135, 0.75, nslices=7, border=F, gradient="y", col=rev(c("#d53e4f", "#f46d43","#fdae61", "#fee08b", "#e6f598", "#abdda4", "#ddf1da")))
text(x=rep(0.118, 7), y=seq(0.28, 0.72, by=0.07), adj=1, cex=0.8, labels=c("0", "0-1", "1-10", "10-100", "100-500", "500-1000", ">1000"))
text(x=0.135, y=0.82, labels="Cases per\n100,000 people", adj=1, cex=0.85)
title(main="Incidence of Measles in the US", line=1, oma=T, adj=0.21)
dev.off()
# End of script ----------------------------------------------------------------
3. Conclusion
We have covered data prep and plotting heatmaps in R using base graphics and ggplot2. ggplot2 seems more consistent with code structure but the base graphics may be useful when combing multiple graphics in complicated ways. Hacking ggplot2 can be harder than fiddling with base graphics. I have not covered heatmaps with dendrograms because they are only useful in specific situations. Perhaps another useful function for heatmaps in R is pheatmap demonstrated here.



Comments